inGeno
An integrated genome and ortholog viewer (inGeno) is designed for genome sequence comparisons, which has been proven to be powerful, in particular to prokaryotic genomes of close phylogenetic distances. The original purpose of this software is to user-friendly visualize the corresponding relationships between orthologous genes. Step by step, a series of algorithms are implemented and integrated together, thus enable a noise-reducing process, a locus collinear block recognition and a text-mining step, which are helpful for users to extract biological information precisely.
Features
- Dot-analysis and plot the results
- User-friendly comparison interface
- Gene information retrieval.
- Linear regression and heuristic locus collinear blocks (LCB) recognition process
- Pseudo-positive orthologous genes reducing procedure (low-noise control)
- Color clusters re-definition using easy-used control panel.
- Text-mining operations helpful for precisely extracting the gene information.
- A sequence conversion tool which can transform rich-format genome sequence files into proteome multi-FASTA format files, which is suitable for making a BLAST or Smith-Waterman analysis.
- Paracel GeneMatcher alignment report is supported, which make sense to obtain ideal sensitive results rapidly.
Documents
| Download |
Program
| Exe:Download | JarRAR :Download | |
| | Bin: Download | JarRAR: Download |
| | Zip: Download | |
| | bin: Download |
(JarRAR file is a package file suitable for most users, users could unpack it, then execute it by ingeno.bat or ./ingeno)
Sourcecode
The Java sourcecode of inGeno is freely available here.
| Development version Download for Linux , (Java JRE 6 required) |
| Screenshots |
Here are some screenshots of the inGeno, which illustrate some features and applications useful for studing genome information, such as linear conservation, rearrangement events, phylogenetic evolution, specific functions which are necessary for reconstructing the metabolism. All the figures are taken from inGeno Ver 0.5a.
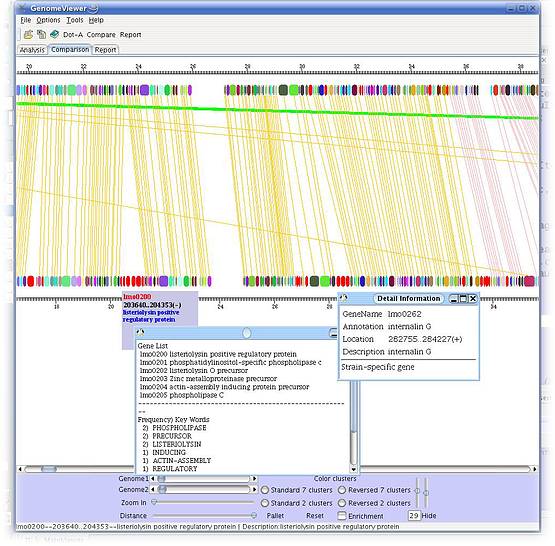
Figure 1. Typically investigation grahpic user interface (GUI). Detail description please read supplemenary materials. Analysis Example ScreenshotsI. Visualization a strain-specific gene islands, which enable a novel metabolism pathway. These genes are continuous in genome, which might to form an operon (Potential operon detection sample (a strain-specific gene island), click to view large figure
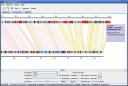
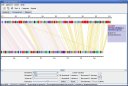
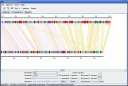
II. Genome rearrangment events investigation samples (green lines in figure), LCB defined by color clusters.