JANE
JANE is written for transcriptome studies on S. aureus. It provides a feasible plan for rapid analysis of EST sequences using a related genome as template to align all the ESTs. The homology modeling results in a virtual genome, with annotation for systematical analysis.
JANA is accessible at http://jane.bioapps.biozentrum.uni-wuerzburg.de
Introduction
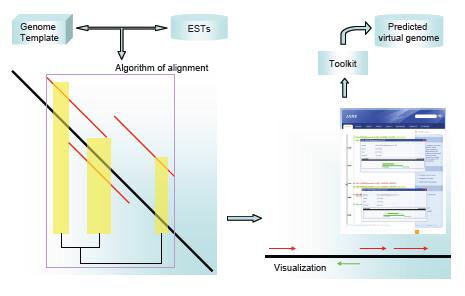
The working flowchart is demontrated in the following figure, Users upload the homologous genome template and sequenced ESTs into Jane software. A heuristic alignment algorithm will compare the sequences and provide an interactive visualization in the browser. With help of the incorporated toolkit, the annotation information can be readily obtained and the sequence can be eventually exported as part of the predicted virtual genome for further analysis.
The alogorithm considers the low-score of alignment region besides the typical HSPs, so that the est2genome alignment benefits the sensitivity from this procedure, whereas most of the classical alignment programs ignore the information. Moreover, the potential direction of alignment segments is particularly taken account of, which helps to find a larger scale of alignment region.
JANE Tutorial
JANE Tutorial
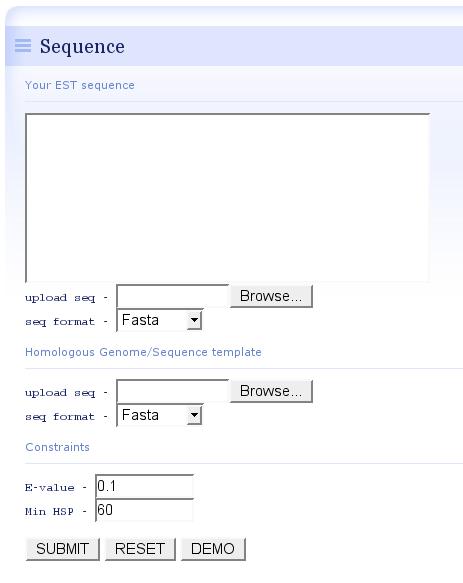
- In the input page, users could upload the EST sequences and homologoous genome template, or simply click the "DEMO" button to accquire a rapid impression of est-2-genome alignment, in this case, 311 ESTs from Blattabacterium sp. (simbiont from the cockroach Blattella germanica) were mapped using the closely-related template genome of Gramella forsetii.
- Alignment result retrievement, the upper figure shows the mapping locations. The alignment figure can be either viewed using mouse hovering the corresponding region or in the lower tables. Moreover, users could generate the virtual genome using the button in the bottom

- The integrated anaylsis toolkit