LS-MIDA
Least Square Mass Isotopomers Distribution Analyzer (LS-MIDA) is an application developed to estimate Mass Isotopomers Distribution from Spectral data by analyzing each peak of given mass and each mass atom fragment.
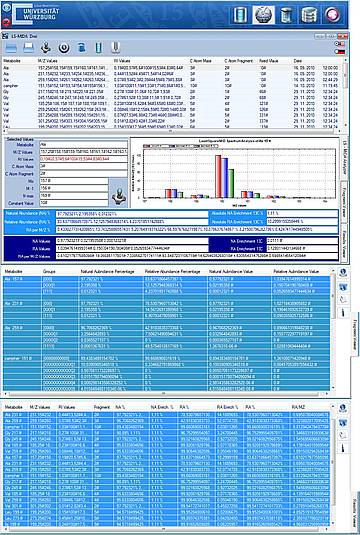
LS-MIDA Drei is the newest version capable of analyzing (isotopic) labeled (Carbon) metabolite based experimental raw data (different amino acids). It estimates Mass Values (minimum and maximum), predicts theoretical Values for pure compounds, draws linear relationships between masses within the range of a compound and performs linear regression analysis to predict Relative Intensity Values with respect to the each Mass to charge Ratio. To give better understanding of obtained results, LD-MIDA Drei visualizes obtained results by drawing a Spectrometer based on mass to charge ratio values, actual relative intensity values, estimated relative intensity values and percentage of relative intensity at each mass to charge ratio. (see Figure 1)
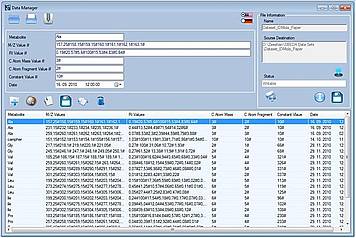
LS-MIDA Drei provides a special graphical user interface for file based experimental raw data manipulation and management which is capable of not only managing user input experimental raw data but also providing options to manage resultant data (output of LS-MIDA Drei). It allows user to create new data files, manage created data files, merge new or already made data files into one or more new or already created files data files and manipulate entries data files. It is an independent file based data management system, doesn't require any external or third party database to install and use. (see Figure 2)
LS-MIDA is implemented in Microsoft Visual Studio Dot Net (C#), integrating formal unified modelling language to scheme from different perspectives and incorporating human computer interaction guidelines, principles and patterns.
LS-MIDA is freely available for academic use with open license. Please write a request at provided contacts.
Related Publications:
- Ahmed Z., Saman Z., Huber C., Hensel M., Schomburg D., Münch R., Eisenreich. W, and Dandekar T. Software LS-MIDA for Efficient Mass Isotopomer Distribution Analysis in Metabolic Modelling. BMC Bioinformatics, 14:218, 2013.
- Ahmed Z., Saman M. and Dandekar T. Unified Modeling and HCI Mockup Designing towards MIDA. Int. Jr. Emerg. Sci., 2:3, 361-382, 2012.
Contact Person:
![]() Prof. Dr. Thomas Dandekar
Prof. Dr. Thomas Dandekar
Dr. Zeeshan AHMED
Scientific Software Engineer
Prof. Thomas Dandekar Group;
Funct. Genomics & Systems Biology.
Department of Bioinformatics;
Biocenter, University of Wuerzburg.
Am Hubland 97074,
Wuerzburg Germany.