Research
DNA repair
DNA is constantly damaged by endogenous and exogenous sources and it has been shown that 80 to 90% of all human cancers are ultimately due to DNA damage. Organisms thus require efficient DNA repair systems to maintain their genomes in a functional state. Nucleotide excision repair (NER) recognizes structurally highly diverse damages, and it is our goal to obtain a general understanding of the sequential process of this repair pathway and exploit its role in cancer therapy. RecQ helicases assume key functions to maintain genomic integrity and are frequently upregulated in many cancers. We investigate the intricate molecular network of RecQ4 to decipher its function and to assess it as a target for cancer therapy.
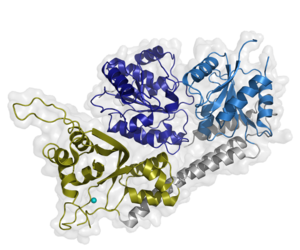
One major player in the NER cascade is the general transcription factor TFIIH. TFIIH is a multiprotein complex that can be divided into core TFIIH (XPB, XPD, p62, p52, p44, p34, and p8) and the cyclin activating kinase complex (CAK: consisting of CDK7, MAT1, and Cyclin H). It is our aim to decipher the intricate network among these subunits. Our data suggest the presence of a redundant interaction network within core-TFIIH, which may serve to minimize the susceptibility to mutational impairment leading to the hallmark nucleotide excision repair syndromes xeroderma pigmentosum or trichothiodystrophy.
RecQ4 is one of five human RecQ helicases that are fundamental for genome maintenance. We solved the structure of the core RecQ4 helicase unit, revealing unique structural domains that are absent in other RecQ proteins. Combined with our functional analysis, we suggest that RecQ4 may employ a helicase mechanism that is different from that of all other human RecQ family members. Our results set the stage to examine the molecular basis of unique RecQ4 genome maintenance functions and analyse the structure-function relationship of patient mutations leading to RecQ4 associated human diseases.
Structure and function of Deubiquitylases
The covalent attachment of ubiquitin by the ubiquitylation cascade (E1, E2, E3) is an important posttranslational protein modification which is involved in the regulation of the majority of cellular pathways in eukaryotes. Deubiquitylases (DUBs) are proteases, that remove ubiquitin from their target proteins and the fine-tuned interplay of both antagonistic reactions permits a rapid and precise response to internal or external stimuli, thereby ultimately determining the fate of the cell.
We focus on the structural and functional characterization of pro- and eukaryotic DUBs with the aim to mechanistically understand their involvement in the development and progression of pathological conditions and ultimately to facilitate the development of pharmacological treatments.
Cancer promoting DUBs: USP25 and USP28
USP25 and USP28 are two closely related DUBs of the large USP family. Both enzymes play important roles in the progression of different types of cancer. However, their roles in healthy tissue such as the control of insulin dependent glucose uptake and the innate immune response (USP25) or DNA-damage signaling (USP28) makes specific inhibition highly desirable.
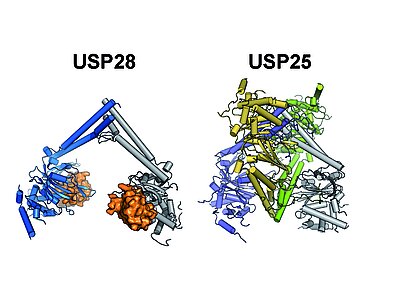
Our crystal-structures of the central USP25 and USP28 catalytic domains show that both share a highly similar architecture, with a novel sub-domain (UCID) that is incorporated into the conserved USP core domain. Our analysis revealed that the UCID mediates protein oligomerization, resulting in the formation of USP28 dimers and USP25 tetramers, with the latter being composed of two interlinked USP28-like dimers. Structure guided functional studies in vitro and in vivo in collaboration with the Popov lab indicate that the tetrameric form of USP25 represents an autoinhibited state and that mutations found in tumors can prevent autoinhibition, resulting in a permanently active enzyme. These data suggest that, in contrast to USP28, which appears to be constitutively active, USP25 activity is regulated by a (currently unknown) upstream factor, thus providing a potential lead to USP25 specific activity modulation. Press release & publication
Both USP25 and USP28 contain a large region of unknown structure located C-terminally to their catalytic domains. In USP25, this region harbors several substrate binding sites while the function of the corresponding area in USP28 is currently unknown.
In order to investigate their structure and create a basis for future functional studies we also expressed and purified the isolated C-terminal regions of both proteins. SEC-MALS analysis and CD-spectroscopy indicated that both exist as monomeric proteins with a mainly alpha-helical fold. We were able to solve the crystal structure of the USP25 C-terminal domain (aa 765-1055) revealing a novel type of protein-interaction domain composed of two sub-domains.
Chlamydial DUBs
Chlamydia trachomatis is an obligate intracellular prokaryote, that is worldwide the major cause of sexually transmitted disease and eye infections. During the majority of its life cycle, it exists within a membrane bound vacuole, the inclusion, and secretes a number of effector proteins to manipulate the host cell’s metabolism to create an intracellular environment suitable for growth and reproduction.
It has been shown that ChlaDUB1, a secreted deubiquitylase that is embedded in the inclusion membrane is responsible for the stabilization of the anti-apoptotic Bcl-2 protein Mcl-1, thereby prolonging the lifespan of the infected cell. This makes ClaDUB1 an attractive drug target for the development of small molecule inhibitors for the treatment of chlamydial infections.
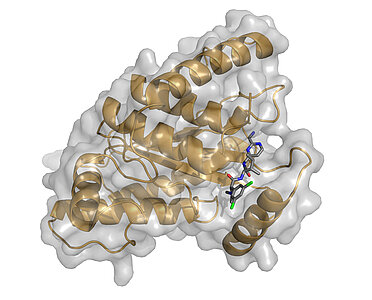
Based on our crystal structure of the ChlaDUB1 protease domain in its apo- and Ub-substrate bound forms, we utilized the resemblance to other (non-DUB specific) Cys-proteases of the CE-clan and employed a ‘target-hopping’ approach. In collaboration with Novartis we identified several small molecules originally designed as inhibitors against the adenoviral protease Adenain as ChlaDUB1 inhibitors with moderate affinity (publication). We are now using these compounds as lead molecules for the development of new, highly specific inhibitors in collaboration with the Sotriffer, Decker and Rudel groups.