P 5: C. Sharma
Virulence factors and regulators required during Campylobacter jejuni infections
State of the art
The Gram-negative Epsilonproteobacterium Campylobacter jejuni is not only the most common cause of bacterial gastroenteritis in humans to date, but is also associated with the development of several secondary autoimmune disorders. In contrast, Campylobacter is considered a commensal in chicken, which are one of the major food-related infection sources. So far it is still unclear which factors are important for its pathogenesis and what triggers the disease development in humans compared to the commensal lifestyle in animals. Campylobacter jejuni lacks many classical virulence factors such as a type III or type VI secretion system known from other enteric pathogens. Therefore, it has been suggested that mainly the metabolic capabilities and motility are required for virulence of these bacteria. Infection studies with Campylobacter have mainly employed in vitro cell culture models using epithelial cancer cell lines, which are limited in their ability to reflect the in vivo situation in the human host and to model the complexity of an intact 3D tissue. Moreover, current mice and chicken animal infection models cannot reflect the human disease development.
Previous Work
The Sharma group has been investigating the mechanisms and functions of small regulatory RNAs (sRNAs) in the pathogenic Epsilonproteobacteria [1, 2], Campylobacter jejuni and Helicobacter pylori. We recently applied a comparative transcriptome analysis to multiple C. jejuni strains to understand how transcriptome differences could contribute to phenotypic differences among strains [3]. This approach is based on differential RNA-sequencing (dRNA-seq) [4] which we had developed for primary transcriptome analysis of H. pylori [5] and our bioinformatics tools for RNA-seq analysis [6]. The comparative dRNA-seq revealed genome-wide transcriptional start site maps in the four C. jejuni strains including strain-specific transcriptional output based on point mutations in promoter regions. It also revealed strain-specific sRNA repertoires which might underlie strain-specific regulation and colonization of different hosts. Now we are investigating the mechanisms and functions of sRNAs in stress response of C. jejuni and are especially interested in their roles in pathogenesis. However, the study of virulence factors is currently hampered by the lack of a suitable infection model.
In a collaborative project between the groups of C. Sharma and H. Walles/M. Metzger that received start-up funding by the Interdisciplinary Center for Clinical Research (IZKF) we have started to establish C. jejuni infections using a previously developed small intestine model (SIM) [7] to show in a proof-of-principle that these 3D tissue models can be used to study bacterial infections. This static SIM is based on a decellularized small intestinal scaffold (SISmuc) that is reseeded with Caco-2 cells in a cell crown.
We have successfully set-up first infections of this SIM as well as methods to track the bacteria during the infection process. Elementary first steps included the determination of cell numbers per cell crown at a given time-point to ascertain an accurate multiplicity of infection (MOI). We have begun to set up suitable readouts to monitor bacterial growth in the 3D tissue models (e.g. CFU counts) and for the infection process such as the disruption of tight junctions (e.g. immunohistochemical stainings and FITC-dextran permeability assays). We have started to further improve the SIM model by addition of a mucus layer and reseeding the SISmuc with the mucus producing E12 cell line. The possibility to study infections of such 3D microenvironments is especially important for extracellular pathogens such as C. jejuni, which thrive in the intestinal mucus layer. In addition, the lab of H. Walles/M. Metzger has started to build tissue models based on human, intestinal primary cells from biopsy samples.
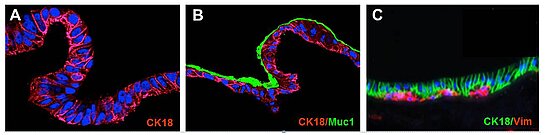
Figure: SISMUC reseeded with human colon epithelial Caco-2 (A) or the mucus producing E12 cells (B). A primary cell co-culture test system is shown in (C). Immunohistological staining: Cytokeratin-18 (CK18), Vimentin (Vim) and Mucin1 (Muc1). Nuclei are stained with DAPI (blue). After 28 days under semi-dynamic culture conditions, E12 cells produce a thick adherent mucus layer (B, green).
Work Plan
In this project we aim to establish human 3D in vitro tissue models to study host-pathogen interactions during C. jejuni infections. We will employ different infection conditions and analyze the infection outcome and translocation of C. jejuni through the polarized epithelial layer using, e.g., invasion assays, histology, ELISA, immuno- and fluorescence microscopy, and Raman Spectroscopy. We will set-up infections with the improved small intestine model including the mucus producing cell lines and models build from primary intestinal cells. After establishing infections on these improved 3D tissue models, we will monitor transcriptome changes of host and pathogen using RNA-seq methods during the course of infection to identify genes which are relevant during infection. Analysis of deletion mutants in the 3D models will then be used to characterize novel virulence genes.
Moreover, we will examine localization of novel virulence factors such as potential adhesins or secreted proteins using super-resolution microscopy and perform 3D live-imaging of the infected tissue models using multi-photon laser-scanning and multiparameter light-sheet fluorescence microscopy. Furthermore, we will infect the 3D tissue models with a bacterial transposon mutant pool generated and globally identify transposon insertions by deep sequencing (Tn-seq). Comparison of transposon insertions distributions in bacteria grown in batch culture with bacteria isolated from infection samples will reveal novel virulence factors and sRNA regulators based on genomic regions depleted for transposon hits, indicating they essentiality for infection. The new models will be useful for infections with other pathogens and will also help to develop a new stomach model for the related gastric pathogen, H. pylori.
References
- Pernitzsch SR & Sharma CM (2012) Transcriptome complexity and riboregulation in the human pathogen Helicobacter pylori. Front Cell Infect Microbiol 2, 14. PubMed
- Pernitzsch et al. (2014) A variable homopolymeric G-repeat defines small RNA-mediated posttranscriptional regulation of a chemotaxis receptor in Helicobacter pylori. PNAS 111, E501-10. PubMed
- Dugar et al. (2013) High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple Campylobacter jejuni Isolates. PLoS Genet 9, e1003495. PubMed
- Sharma CM & Vogel J (2014) Differential RNA-seq: the approach behind and the biological insight gained. Curr Opin Microbiol 19C, 97-105. PubMed
- Sharma CM et al. (2010) The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250-5. PubMed
- Förstner et al. (2014) READemption-a tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics 30(23), 3421-3. PubMed
- Pusch et al. (2011) The physiological performance of a three-dimensional model that mimics the microenvironment of the small intestine. Biomaterials 32, 7469-78. PubMed