P 2: T. Rudel
Host factors required for the initiation and propagation of Chlamydia trachomatis infections
State of the art
The human pathogen Chlamydia trachomatis causes trachoma and sexually transmitted diseases in hundreds of millions patients annually. Chlamydia grow in a membrane bound vacuole (“inclusion”) inside the host cell. In a complex developmental cycle, infectious non-replicating elementary bodies (EB) start infection and transform to non-infectious replicating reticulate bodies (RB) that after several rounds of multiplication transform back to EB and leave the host cell. The obligate intracellular lifestyle of Chlamydia has hampered investigation of pathogenicity mechanisms in this pathogen, since bacteria do not grow outside of host cells.
Human cancer cell lines have mainly been used as infection models and for bacterial propagation. Innate immune signaling and metabolic pathways are altered in cancer cells, therefore, basic processes of Chlamydia infection cannot be investigated in transformed cells. As no animal model is available which mimics natural infection with C. trachomatis, the mouse pathogen C. muridarum has been established as in vivo infection model for C. trachomatis. However, our recent data show that primary human and mouse cells fundamentally differ in Chlamydia infection responses, limiting the relevance of the mouse model [1].
Previous Work
The Rudel group has been investigating the innate immune response elicited by infection with Chlamydia. We demonstrated that the pathogen interferes with apoptotic death of the host cell to avoid premature interruption of the developmental cycle and consequently eradication [2-4]. We recently identified the receptor tyrosine kinase EphA2 as adherence and entry receptor of EB which induces the rapid and long-lasting activation of the PI3 kinase pathway [5]. Active PI3 kinase is absolutely required for keeping apoptosis resistance in the infected cell, e.g. by upregulation of the apoptosis inhibitor Mcl-1 [6]. Mcl-1 is additionally stabilized by the chlamydial deubiquitinase ChlaDUB1 that is secreted into the inclusion membrane and stabilizes Mcl-1 by deubiquitination. We have experimental evidence that ChlaDUB1 has other targets, which are required for the normal development of Chlamydia.
In a combined proteome and whole genome RNA interference approach, we have already defined the set of human proteins associated with the chlamydial inclusion that are required for chlamydial replication and development. In addition, tools for genetic manipulation of Chlamydia (transgene expression, knock in, inducible gene expression) have been established that can be used to investigate the role of secreted chlamydial proteins in the adaptation in primary human cells.
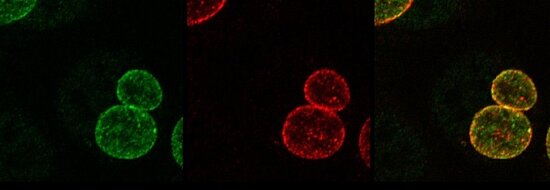
Figure: Secretion of chlamydial ChlaDUB1 (red) into the inclusion membrane (green) in primary fimbriae cells
The Dandekar group are experts in modeling interactions of bacteria and viruses with human cells [7-12]. We provide for each step bioinformatics expertise so that both regulatory [7,10,12] and metabolic aspects [8,11] of the Chlamydia infection can be modelled as well as co-infection with N. gonorrhoeae including different host cell types and responses (primary polarized fallopian and cervical cells, cancer cells). Apoptotic processes and their modulation are modelled with high resolution [10]. Boolean models capture decision processes in innate immunity and host cell responses. Dynamical modelling includes semi-quantitative models and ODE-based detailed descriptions of cellular switches [9]. Large-scale metabolic models will compare host and bacterial metabolism [7,11]. Pathogen specific metabolism and regulation will be modelled building on our experience of analyzing large-scale transcriptome datasets [7,8].
Work Plan
Architecture, physiologic and metabolic state of primary human polarized cervical and fallopian tube cells deliver a new model substantially different from cancer cell infection models used for Chlamydia in the past. Thomas Rudel´s group will establish an engineered vascularized infection model from cervical and fallopian tube epithelial cells. Since N. gonorrhoeae and Chlamydia frequently cause co-infections in patients and infect the same tissue, this infection model will be developed together with the group of Vera Kozjak-Pavlovic (project 3). Gene regulation in the infection model will be investigated by dual RNA-Seq. Raman Spectroscopy will be used to follow up the cross-talk of Chlamydia with primary host cells. A focus will be the innate immune response and metabolic reprogramming of host cells that allows Chlamydia to establish a successful infection.
Thomas Dandekar will model this, starting from transcriptome and metabolome data. Furthermore, in a combined proteome and whole genome RNA interference approach, the Rudel group has already defined the set of human proteins that are required for chlamydial replication and development. Those genes which are essential for chlamydial development and are regulated during infection of primary cells will be investigated by both groups combining modelling and experiment. Particularly for obligate intracellular bacteria that depend on the metabolites from their host cell for generation of progeny, the response of a non-replicating polarized host cell has to be orchestrated by the incoming pathogen to ensure the uptake of the EB, formation of the inclusion and the provision of nutrients during the replicative phase. In addition, innate immune response has to be directed towards the survival of the host at least until the developmental cycle of Chlamydia has been completed.
References
- Siegl et al. (2014). Tumor Suppressor p53 Alters Host Cell Metabolism to Limit Chlamydia trachomatis Infection. Cell Rep 9, 918-929. PubMed
- Sharma et al. (2011). HIF-1alpha is involved in mediating apoptosis resistance to Chlamydia trachomatis-infected cells. Cellular microbiology 13, 1573-1585. PubMed
- Rudel et al. (2010). Interactions between bacterial pathogens and mitochondrial cell death pathways. Nature Reviews Microbiology 8, 693-705. PubMed
- Bohme et al. (2010). Chlamydia trachomatis-infected host cells resist dsRNA-induced apoptosis. Cell Microbiol 12, 1340-1351. PubMed
- Subbarayal et al. EphrinA2 receptor (EphA2) is an Invasion and Intracellular Signaling receptor for Chlamydia trachomatis. PLoS pathogens 23;11(4):e1004846. PubMed
- Rajalingam et al. (2008). Mcl-1 is a key regulator of apoptosis resistance in Chlamydia trachomatis infected cells. PLOS One 3, e3102. PubMed
- Cecil et al. (2014) Modelling antibiotic and cytotoxic isoquinoline effects in Staphylococcus aureus, Staphylococcus epidermidis and mammalian cells. Int J Med Microbiol. 305(1):96-109. PubMed
- Cecil et al. (2014) Antigen profiling analysis of vaccinia virus injected canine tumors: Oncolytic virus efficiency predicted by boolean models. Bioengineered 5(5):319-25. PubMed
- Mischnik et al. (2014) A comparative analysis of the bistability switch for platelet aggregation by logic ODE based dynamical modeling. Mol Biosyst. 10, 2082-2089. PubMed
- Fricke et al. (2014) Quantitative single-molecule localization microscopy combined with rule-based modeling reveals ligand-induced TNF-R1 reorganization toward higher-order oligomers. Histochem Cell Biol. 142,91-101. PubMed
- Winstel et al. (2013) Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nature Commun. 4,2345. PubMed
- Audretsch et al. (2013) A semi-quantitative model of Quorum-Sensing in Staphylococcus aureus, approved by microarray meta-analyses and tested by mutation studies. Mol. BioSyst 9,2665-80. PubMed