P 10: S. Bartfeld
Analysis of the innate immune response of gastrointestinal epithelium in 3D organoids
State of the art
The epithelial lining of the host's gastrointestinal tract is the prime interface for interactions with pathogens and acts as physical and immunological barrier between the gut microbiota and the body. A central function of the gastrointestinal epithelium is the establishment and maintenance of host-microbe homeostasis thereby minimizing any inflammation by commensals, yet still enabling the immediate sensing of pathogens and initiating a strong inflammatory response to infections. The molecular components generally responsible for this sensing are the pattern recognition receptors (PRRs) and their downstream signaling cascades. PRRs recognize microbe or pathogen-associated molecular patterns (MAMPs or PAMPs). While the general scheme of PRR-MAMP signaling is quite well understood, many questions remain to be answered. For example, there have been substantial discrepancies in the description of the presence and functionality of the components of the innate immune signaling cascade in the gastrointestinal tract in mouse and human. Studies using various available gastric cancer cell lines or primary material arrive at very different conclusions (summarized in Abreu 2009). It is likely, that results are so diverse because cell lines have accumulated many mutations (because they have been generated from cancer and acquired additional mutations over decades in culture) and thus the signaling cascades may be altered in comparison to the primary epithelium. The evaluation of the functionality of PRR signaling cascades on primary epithelium has so far been hampered by limited availability of primary material.
Previous work
Recent advances in the field of stem cell biology have now provided the ideal culture system to address these questions. This culture system allows theoretically endless culture of primary cells from virtually every human patient, as well as from laboratory animals. In this approach, epithelial stem cells are isolated from the respective organ, placed in an extracellular Matrix (Matrigel) and supplemented with an organ-specific cocktail of growth factors and inhibitors. The stem cells subsequently divide and grow into 3-dimensional mini-versions of the organ they have been generated from, so called "organoids". Organoids are genetically stable and faithfully represent the tissue they originate from (reviewed in Bartfeld and Clevers, 2017). Importantly, organoids are amenable to a wide range of laboratory techniques including (live and confocal) microscopy, RNA and protein analysis and can also be modified using lentivirus or CRISPR/Cas9. Organoids have been used to study infection with several pathogens (reviewed in Bartfeld 2016). For example, human gastric organoids can be infected with H. pylori and it has been shown that the innate immune response to infection depends on the differentiation of cells in the organoids (Bartfeld et al 2015).
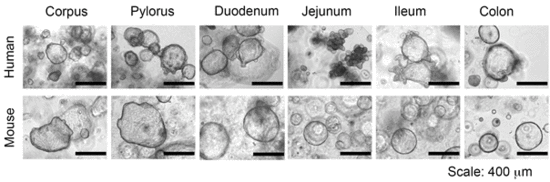
Figure: Human and murine gastrointestinal organoids.
Work Plan
In this study, we aim to generate a biobank of gastrointestinal organoids and profile the organoids for their PRR signaling components. For this, PCR will be used to verify the expression of single genes, especially the components of the innate immune signaling cascade (PRR/NF-kappaB signaling). To examine, whether stimulation of expressed PRRs lead to activation of the signaling cascade, expression of target genes in response to purified PRR-ligands will be examined using qRT-PCR. After identification of functional differences in PRR signaling, their importance for infection, for example for the gastric pathogen Helicobacter pylori will be analyzed. Analysis of mutants will be used to further characterize specific signaling components.
We will then investigate the molecular basis for potential specific regulations of PRR signaling components. Mining the literature and available transcriptomic data of organoids, candidate genes that are implicated in the regulation of PRR signaling will be identified. We will analyze the function of candidate genes by comparing organoids from knockout mice (if they exist) or generate organoids with CRISPR/Cas9 mediated knockout and lentiviral overexpression. Overall, this project will generate an "atlas" of PRR signaling throughout the mouse and human gut, identify the impact of PRR signaling on the host response to H. pylori, and analyze the mechanics of PRR regulation in the gut.
References
- Abreu MT: Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010 Feb;10(2):131-44. doi: 10.1038/nri2707. PubMed
- Bartfeld S: Modeling infectious diseases and host-microbe interactions in gastrointestinal organoids. Dev Biol. 2016 Dec 15;420(2):262-270. doi: 10.1016/j.ydbio.2016.09.014. PubMed
- Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, Vries R, Peters PJ & Clevers H: In Vitro Expansion of Human Gastric Epithelial Stem Cells and Their Responses to Bacterial Infection. Gastroenterology. 2015 Jan;148(1):126-136.e6. doi: 10.1053/j.gastro.2014.09.042. PubMed
- Bartfeld S, Clevers H: Stem cell-derived organoids and their application for medical research and patient treatment. J Mol Med (Berl). 2017 Jul;95(7):729-738. doi: 10.1007/s00109-017-1531-7. PubMed