Group Spahn - Nanoscale Bacteriology
Welcome to research group "Nanoscale Bacteriology"!
The Nanoscale Bacteriology Lab investigates the molecular organization of bacterial cells. We employ diverse microscopy methods, in particluar super-resolution microscopy. This set of microscopy methods allows to visualize biological specimen and structures with a spatial resolution of a few nanometers. Compared to conventional, diffraction-limited microscopy (e.g. widefield or confocal fluorescence microscopy), this represents a resolution increase by a factor of 10 - 30. With this greatly enhanced resolution, we are now able to study proteins, nucleic acids and other biomolecules on the near-molecular scale an in the context of individual, intact cells.
The methods or workflows for multicolor super-resolution imaging in bacteria is nowadays still limited. We thus establish and develop methods that allow the simultaneous visualization of different target molecules. This includes single-molecule based techniques (single-molecule localization microscopy, SMLM), stimulated emission depletion (STED) microscopy and live-cell imaging. Together with artificial intelligence, these imaging methods can provide rich information on diverse biological questions. We are also interested in the development of smart microscopy approaches, which can for example accelerate drug screening at high spatial resolution.
Our biological focus lies on the investigation of the transertion process in gram-negative bacteria. Transertion describes the coupling of transcription, translation and insertion of membrane proteins, which represents a postulated, but still insufficiently studied (an unproven) mechansim that dynamically couples the chromsomal DNA (the so-called nucleoid) to the inner membrane. As bacterial cells are tiny (an E. coli cell in rich medium is ~ 1 µm wide and 3 - 5 µm long), we need high-resolution methods to study the nature of such processes. Using systematic perturbation of cellular processes with drug treatments, we could show that protein biosynthesis strongly contributes to the organization of the E. coli nucleoid. In our ongoing and future work, we want to decipher the molecular mechanisms that mediate bacterial organization, both in our model organism E. coli, as well as in gram-negative pathogens.
Our broad method spectrum allows us to study many more exciting biological questions. This includes:
- Architecture of bacterial biofilms
- Antibiotic resistance mechansims
- Host-pathogen interactions

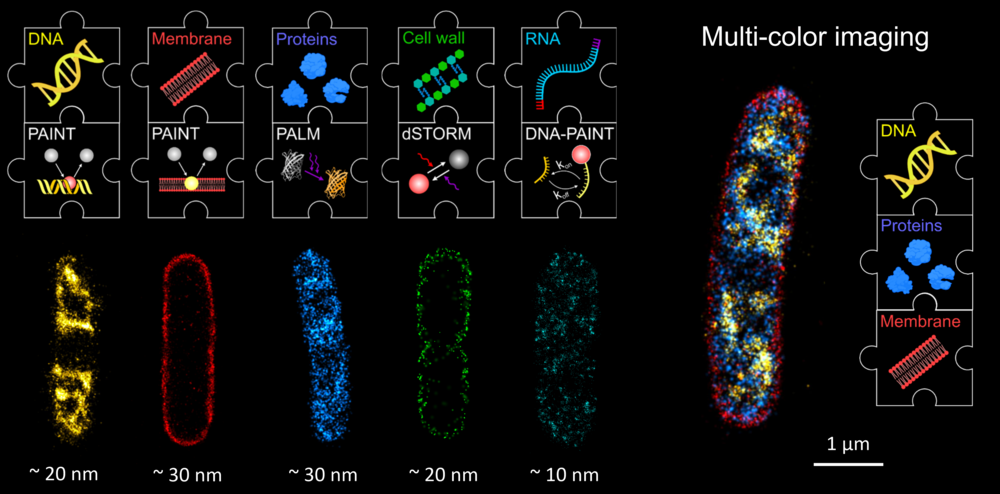
Single-molecule localization microscopy (SMLM)
One pillar of our research is the development of super-resolution approaches, particularly for their use in microbiology. We mainly use single-molecule localization microscopy (SMLM) (Fig. 1), in which temporally separated emitters (single dye molecules) can be localized with high precision, allowing the reconstruction of an artificial image with increased resolution (typically 10 - 40 nm) and contrast.
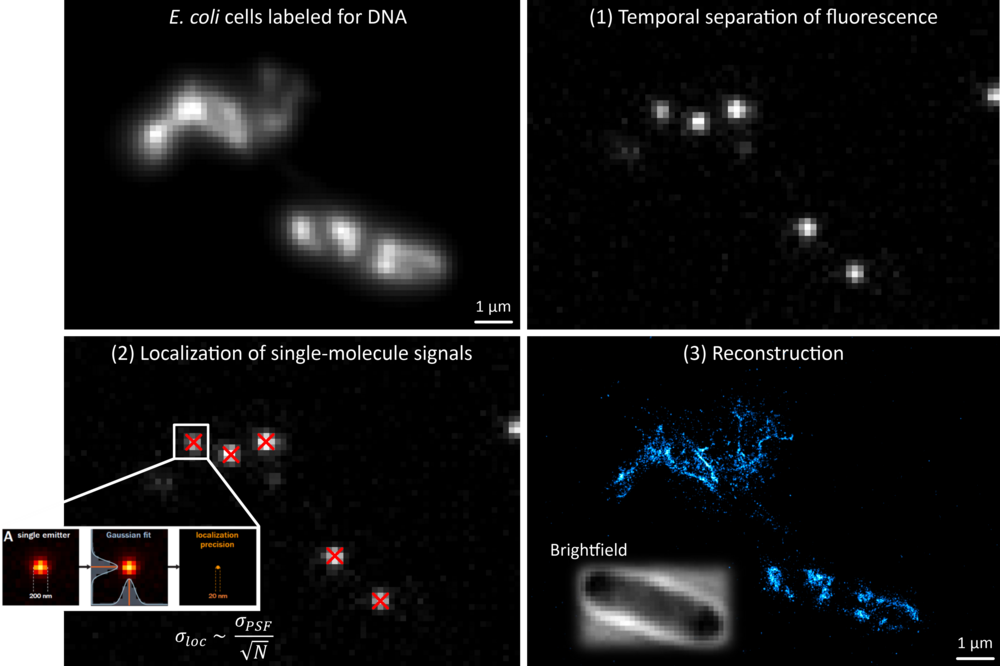
In particular, we developed tools to study the organization of chromosomal DNA (the ‘nucleoid’) in fixed bacterial cells. Starting with an EdU-based metabolic labeling approach and direct Stochastic Optical Reconstruction Microscopy (dSTORM) [1], we switched to transiently binding labels that can be used for Point Accumulation for Imaging in Nanoscale Topography (PAINT) [2]. Together with the lab of Luke Lavis (Janelia Farm, United States), we tested a palette of DNA- and membrane-targeted PAINT probes that enable multiplexed imaging of the nucleoid and membranes in various color-combinations. This simple approach is very versatile and enables super-resolution imaging independent of active replication. We find that the E. coli nucleoid is highly structured during fast growth, and that the nucleoid is positioned in close proximity to the inner membrane [3].
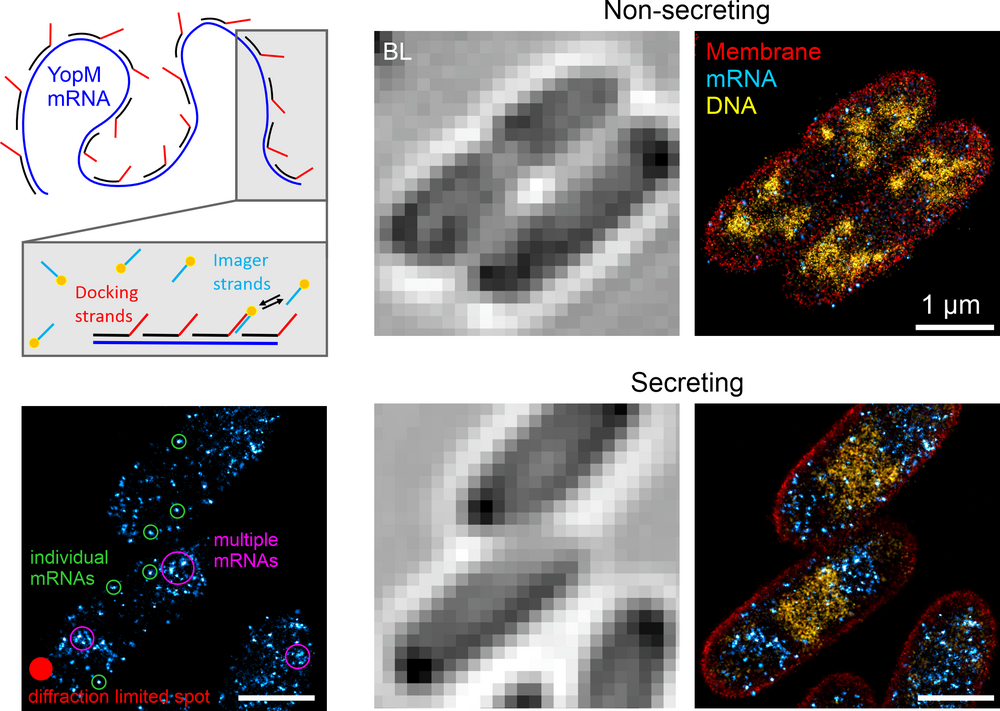
Recently, we used this approach to study the response of the E. coli nucleoid to perturbations. Tracking the time-resolved reaction to antibiotic treatments unraveled a rapid nucleoid reorganization, particularly during inhibition of transcription and translation. We think that this reorganization occurs due to disruption of transertion, the dynamic coupling of transcription, translation and membrane insertion/translocation of proteins [4].
SMLM allows for the visualization of various biological structures (Fig. 3). The combination of different SMLM modalities in multicolor imaging provides a sharp view on the organization of individual bacterial cells.
STED microscopy
To enable high-quality single-molecule imaging, PAINT probes need to be titrated. However, this can be challenging for heterogeneous structures that show both high-density and low-density distributions. As an alternative to SMLM, we explored the use of transient labels at high concentration. A sufficiently high concentration of PAINT probes (typically 100 – 500 nM) provides pseudo-permanent labeling, which can be used for efficient STED imaging [5]. The constant exchange of labels circumvents photobleaching, which is often a limiting factor for high-quality STED imaging.
The optical sectioning capability of STED microscopy navigates the problem of high background using non-fluorogenic probes. We successfully applied the concept of STED with exchangeable labels to IRIS probes such as Lifeact and also fluorescently labeled DNA oligos as used in DNA-PAINT [5, 6]. We think that this concept is generalizable to all labels that fulfill certain kinetic properties (high nM to low µM affinity, high on- and off-rates).
![Fig. 3: xSTED microscopy with exchangable probes. [A] Schematic representation of the concept. Target structures (here: plasma membrane) are labeled pseudo-permanently with a high concentration of transiently binding probes. Bleached molecules are randomly exchanged by new, unbleached probes. [B] 2-color xSTED measurement of <i>E. coli</i> cells stained for DNA (yellow) und membranes (red). [C] xSTED measurement of mitochondria and the ER network. [D] 2-color xSTED measurement of DNA-labeled target molecules (based on DNA-PAINT probes). image description](/fileadmin/_processed_/2/a/csm_xSTED_8741ac0110.png)
Chromosome organization in bacteria
Our biggest biological interest lies in understanding how bacteria organize their chromosomes. We are intrigued by the diversity of shapes that the nucleoid can adapt, raising questions about the underlying mechanismsIt is long known that the nucleoid morphology strongly varies in dependence of external and internal factors, including nutrient availability, temperature, growth phase or the presence of certain compounds (e.g. antibiotics, toxins). As nucleoid size only slightly exceeds the diffraction limit, super-resolution microscopy is ideally suited to study its nanoscale architecture and to unravel subtle differences between the beforementioned conditions (Fig. 4).
For example, some antibiotics have a strong effect on cell and nucleoid morphology (Fig. 4A). This is employed in image-based drug screening, in which the different phenotypes serve as a readout for the coarse mode-of-action (MoA) of a drurg. We think that a strongly improved resolution enables a more detailed and exact assessment of antibiotic MoAs.
In collaboration with Helge Bode (MPI for Terrestrial Microbiology, Marburg) we studied the distribution of the antimicrobial peptide PAX (Peptide Antimicrobial Xenorhabdus) (Fig. 4B) [7]. Super-resolution imaging revealed that the peptides flood the cytosol while also solubilizing the membrane.
Referenzen
[1] Spahn, C., Endesfelder, U. & Heilemann, M. “Super-resolution imaging of Escherichia coli nucleoids reveals highly structured and asymmetric segregation during fast growth.” Journal of Structural Biology 185 (3), 243-249 (2014). [link]
[2] Spahn, C., Glaesmann, M., Grimm, J.B., Ayala, A.X., Lavis, L.D., Heilemann, M., “A toolbox for multiplexed super-resolution imaging of the E. coli nucleoid and membrane using novel PAINT labels.” Scientific Reports 8, 14768 (2018). [link]
[3] Spahn, C., Middlemiss, S., Gómez-de-Mariscal, E., Henriques, R., Bode, H. B., Holden, S., Heilemann, M., “The nucleoid of rapidly growing Escherichia coli localizes close to the inner membrane and is organized by transcription, translation, and cell geometry.” Nat. Commun. 16, 3732 (2025) [link]
[4] Woldringh, C., “The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation.” Mol. Microbiol. 45, 17-29. [link]
[5] Spahn, C., Grimm, J.B., Lavis, L.D., Lampe, M., Heilemann, M., “Whole-Cell, 3D, and Multicolor STED Imaging with Exchangeable Fluorophores.” Nano Letters 19 (1), pp 500–505 (2018). [link]
[6] Spahn, C., Hurter, F., Glaesmann, M., Karathanasis, C., Lampe, M., Heilemann, M., “Protein-Specific, Multicolor and 3D STED Imaging in Cells with DNA-Labeled Antibodies.” Angewandte Chemie 58(52), pp 18835-18838 (2019). [link]
[7] Vo T.D., Spahn, C., Heilemann, M., Bode, H.B., “Microbial cationic peptides as a natural defense mechanism against insect antimicrobial peptides.” ACS Chemical Biology 16(3), pp 447-451 (2021) [link]
- Ermoli, F. #, Spahn, C.#, Sadhir, I., Bode, H.B., Diepold, A., "Activation of the Yersinia type III secretion system induces large-scale spatial reorganization of chromosomal and virulence plasmid DNA." Cell Reports, accepted (2025)
- Ermoli, F., Malengo, G., Spahn, C., Brianceau, C., Glatter, T., Diepold, A.,"Yersinia actively downregulates type III secretion and adhesion at higher cell densities." PLoS Pathog. 21(8):e1013423 (2025) [link]
- Spahn, C., Middlemiss, S., Gómez-de-Mariscal, E., Henriques, R., Bode, H. B., Holden, S., Heilemann, M., “The nucleoid of rapidly growing Escherichia coli localizes close to the inner membrane and is organized by transcription, translation, and cell geometry.” Nat. Commun. 16, 3732 (2025) [link]
- Pandi, A., Adam, D., Zare, A., Trinh, V.T., Schaefer, S.L., Wiegand, M., Klabunde B., Bobkova, E., Kushwaha, M., Foroughijabbari, Y., Braun, P., Spahn, C., Preußer, C., von Strandmann, E.P., Bode, H.B., von Buttlar, H., Bertrams, W., Jung, A.L., Abendroth, F., Schmeck, B., Hummer, G., Vázquez, O., Erb, T.J. “Cell-free biosynthesis combined with deep learning accelerates de novo-development of antimicrobial peptides.” Nat. Commun. 14, 7197 (2023) [link]
- Spahn, C., Gómez-de-Mariscal, E., Laine, F.L., Pereira, P. M., von Chamier, L., Condiut, M., Pinho, M.G., Jacquemet, G., Holden, S., Heilemann, M. & Henriques, R., “DeepBacs for multi-task bacterial image analysis using open-source deep learning approaches.” Commun. Biol. 5, 688 (2022) [link]
- Koller, N., Höllthaler, P., Barends, M., Döring, M, Spahn, C., Durán, V., Costa, B., Becker, J., Heilemann, M., Kalinke, U., Tampé, R., “Nanoscale organization of the MHC I peptide-loading complex in human dendritic cells.” Cell. Mol. Life Sci. 79(9):477 (2022) [link]
- Chamier, v.L.#, Laine#, R.F., Jukkala, J., Spahn, C., Krentzel, D., Nehme, E., Lerche, M., Hernández-Pérez, S., Mattila, P.K., Karinou, E., Holden, S., Solak, A.C., Krull, A., Buchholz, T.-O., Jones, M.L., Royer, L.A., Leterrier, C., Shechtman, Y., Jug, F., Heilemann, M., Jacquemet, G., Henriques, R., “Democratising deep learning for microscopy with ZeroCostDL4Mic.” Nature Communications 12, 2246 (2021) [link]
- Vo T.D.#, Spahn, C.#, Heilemann, M., Bode, H.B., “Microbial cationic peptides as a natural defense mechanism against insect antimicrobial peptides.” ACS Chemical Biology 16(3), pp 447-451 (2021) [link]
- Glogger, M., Spahn, C., Enderlein, J., Heilemann M., “Multi-Color, Bleaching-Resistant Super-Resolution Optical Fluctuation Imaging with Oligonucleotide-Based Exchangeable Fluorophores.” Angewandte Chemie (2020). [link]
- Spahn, C., Hurter, F., Glaesmann, M., Karathanasis, C., Lampe, M., Heilemann, M., “Protein-Specific, Multicolor and 3D STED Imaging in Cells with DNA-Labeled Antibodies.” Angewandte Chemie 58(52), pp 18835-18838 (2019). [link]
- Spahn, C., Grimm, J.B., Lavis, L.D., Lampe, M., Heilemann, M., “Whole-Cell, 3D, and Multicolor STED Imaging with Exchangeable Fluorophores.” Nano Letters 19 (1), pp 500–505 (2018). [link]
- Spahn, C.#, Glaesmann, M.#, Grimm, J.B., Ayala, A.X., Lavis, L.D., Heilemann, M., “A toolbox for multiplexed super-resolution imaging of the E. coli nucleoid and membrane using novel PAINT labels.” Scientific Reports 8, 14768 (2018). [link]
- Gao, Y.#, Spahn, C.#, Heilemann, M., Kenney, L.J., “The Pearling Transition Provides Evidence of Force-Driven Endosomal Tubulation during Salmonella Infection.” mBio 9, e01083-18 (2018). [link]
- Diekmann, R., Wolfson, D.L., Spahn, C., Heilemann, M., Schüttpelz, M. & Huser, T. “Nanoscopy of bacterial cells immobilized by holographic optical tweezers.” Nature Communications 7, 13711 (2016). [link]
- Spahn, C., Herrmannsdӧrfer, F., Kuner, T. & Heilemann, M. “Temporal accumulation analysis provides simplified artifact-free analysis of membrane-protein nanoclusters.” Nature Methods 13, 963–964 (2016). [link]
- Devraj, K., Poznanovic, S., Spahn, C., Schwall, G., Harter, P., Mittelbronn, M., Antoniello, K., Paganetti, P., Muhs, A., Heilemann, M., Hawkins, R.A., Schrattenholz, A. & Liebner, S. “BACE-1 is expressed in the blood-brain barrier endothelium and is upregulated in a murine model of Alzheimer’s disease.” Journal of Cerebral Blood Flow & Metabolism 36, 1281–1294 (2016). [link]
- Foo, Y. H.#, Spahn, C.#, Zhang, H., Heilemann, M. & Kenney, L. J. “Single cell super-resolution imaging of E. coli OmpR during environmental stress.” Integr. Biology 7, 1297–1308 (2015). [link]
- Spahn, C., Cella-Zannacchi, F., Endesfelder, U. & Heilemann, M. “Correlative super-resolution imaging of RNA polymerase distribution and dynamics, bacterial membrane and chromosomal structure in Escherichia coli.” Methods and Applications in Fluorescence 3, 014005 (2015). [link]
- Raulf, A., Spahn, C., Zessin, P. & Heilemann, M. “Click chemistry facilitates direct labelling and super-resolution imaging of nucleic acids and proteins.” RSC Advances 4, 30462–30466 (2014). [link]
- Spahn, C., Endesfelder, U. & Heilemann, M. “Super-resolution imaging of Escherichia coli nucleoids reveals highly structured and asymmetric segregation during fast growth.” Journal of Structural Biology 185 (3), 243-249 (2014). [link]
- Klehs, K., Spahn, C., Endesfelder, U., Lee, S.F., Fuerstenberg, A. & Heilemann, M. “Increasing the Brightness of Cyanine Fluorophores for Single-Molecule and Superresolution Imaging.” ChemPhysChem 15, 637–641 (2014). [link]
Dr. Christoph Spahn
Josef-Schneider-Straße 2

Kilian Andress
Josef-Schneider-Straße 2

Ruilan Xu
Josef-Schneider-Straße 2
Lars Köhler

PostDoc Position in Infection Biology and Advanced Imaging (f/m/d)
The Nanoscale Bacteriology Lab, led by Dr. Christoph Spahn, is looking for a PostDoc to study the intracellular organization of intracellular pathogens during infection.
Our research focusses on understanding the subcellular organization of bacteria and their response to antimicrobial compounds. We combine super-resolution microscopy (Spahn et al., 2018), drug treatments (Spahn et al., 2025) artificial intelligence (Spahn et al., 2022) and biochemical approaches to understand both fundamental concepts of bacterial cell biology and antibiotic action. Recently, we started to investigate how secretion alters the intracellular organization of pathogens (Ermoli et al., 2025).
In the advertised position, the successful candidate will investigate bacterial secretion during host-pathogen interaction. We will employ a combinatorial approach using live-cell imaging, multicolor super-resolution microscopy and genetics to gain fundamental knowledge about the dynamics of effector protein secretion and the effect on both host cells and pathogens.
The position is scheduled to last until the end of 2028.
Qualification profile
As a PostDoc applicant, you have completed a PhD in life sciences or related fields.
Essential qualifications
- Enthusiasm for science/basic research
- A strong background in the field of life sciences
- Experience in infection biology and/or molecular biology of bacterial pathogens
- Experience in bacterial and mammalian cell culture
- Experience in super-resolution microscopy or other advanced microscopy techniques, as well as image analysis
- Willingness to learn new techniques and explore novel ground
- Excellent team spirit and willingness to teach students
- Fluent English
Optional qualifications
- Experience in programming or with AI
- Experience in biochemistry
What we offer
At Julius-Maximilians-Universität Würzburg, more than 30,000 people from over 100 countries come together to study, research, teach and work. As a central institution of the University, the Rudolf Virchow Center is a modern research center with more than 100 international scientists investigating the molecular causes of health and disease. It has been established as a unique institution for translational bioimaging combining all existing imaging core structures and microscopy facilities.
The Rudolf Virchow Center is a highly competitive international research institute with an outstanding infrastructure. We have in-house access to state-of-the-art instrumentation and to an extensive range of modern biophysical and cell biological equipment. We are integrated into a stimulating cross-disciplinary environment that includes our partners at the Biocenter, the physics department, the Max Planck Research Group in Systems Immunology, the Institute for Molecular Infection Biology (IMIB), Helmholtz Institute for RNA-based Infection Research (HIRI) and the University Hospital in Würzburg.
Würzburg is located in the beautiful, wine-growing area of Franconia, in proximity of Frankfurt (1 hour by train) and Munich (2 hours). We have a lot of sun and many festivals in the summer, as well as impressive historical buildings and recreative nature.
The salary is commensurate with training and experience according to Collective Agreement for the Public Service of German Federal States TV-L (100%).
You will have access to both national and public-service pension schemes (VBL), health care, and are entitled to 30 days of holiday in addition to 13 annual public holidays in Bavaria. Our Welcome Center supports international candidates in finding accommodation as well as in administrative matters - and the University of Würzburg offers support for researchers with children, including flexible working hours and childcare.
Interested?
Applications including a cover letter, a detailed CV (2 pages max), a short summary of past research projects with publications (3 pages max), copies of certificates, and the contact information of two referees, should be sent via email as a single pdf file (not exceeding 10 MB). Applications will be screened continuously until the position is filled.
The University of Würzburg is an equal opportunity employer. As such, we explicitly encourage applications from qualified women.
Please send your application to:
Christoph.spahn@uni-wuerzburg.de with Cc to Inka.robinson@uni-wuerzburg.de
If you want to know more about the individual project and/or have further questions about the position, please contact me via the mail above.


![Fig. 4: Super-resolution imaging of drug-treated <i>E. coli</i> cells. [A] Phenotypes of bacteria that were treated with different antibiotics. [B] Localization and effect of the antimicrobial peptide PAX.](/fileadmin/_processed_/6/7/csm_Drug_treatments_ABs_first_e3614aaac5.png)