New models and better data for macromolecular structure determination
07/08/2019The structures of proteins, RNA and DNA are the key to our understanding of life. These structures are measured with X-ray or neutron diffraction, but the data are rarely perfect. Researchers at the University of Würzburg are now developing a new software to analyse this data.
For over 50 years, X-ray crystallography has been the foremost method for the determination of three dimensional structures of biological macromolecules. The method has revolutionized biochemistry, allowing a fundamental insight into the chemistry of living cells, and enabling the development of new drugs and vaccines.
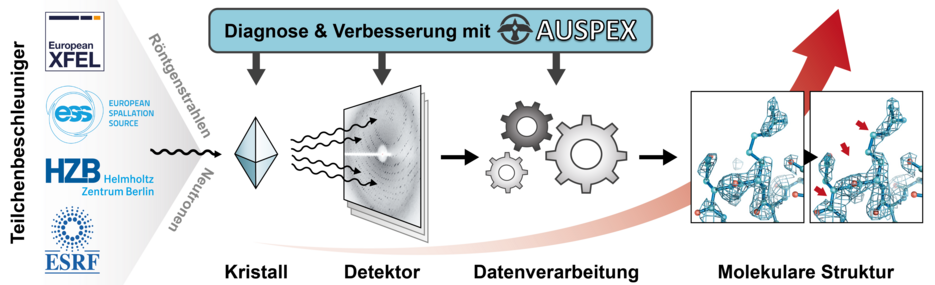
To obtain such a structural model, a crystal (of DNA, for example) is irradiated with neutrons or X-rays, and the diffracted radiation is recorded with a detector. With these diffraction data, a three dimensional model of the molecule is constructed to explain the measured data as well as possible. Consequently, the quality of diffraction data and this knowledge determine the quality of the molecular structure and all biological or medicinal insights it may offer.
In recent years, large sums have been invested for this technology both in Germany and in Europe, including the 3.4 km long European X-Ray Free Electron Laser (EuXFEL) in Hamburg. These innovations drastically increased the speed of measurements and amplified the amount of data available for structure solutions. “However, in order to exploit this wealth of data, we need reliable automatic analysis, that give users direct feedback about their measurements,” stresses Dr. Andrea Thorn.
To tackle this challenge, the junior group leader in the research lab of Prof. Hermann Schindelin at the Rudolf-Virchow Center for Experimental Biomedicine (University of Würzburg) is currently developing a new computer program called AUSPEX (www.auspex.de). This software will help crystallographers to optimize both their measurements as well as data processing. In May of this year, the Federal Ministry for Research and Education (BMBF) agreed to fund this project, which allows Thorn to bring together collaborators from four major European crystallography facilities: the European Free X-ray Laser EuXFEL in Hamburg, the BESSY synchrotron facility in Berlin, the European Synchrotron Radiation Facility in Grenoble (France) and the European Spallation Source (ESS) in Lund (Sweden). The researchers anticipate that new quality standards will be set for diffraction data measured at these (and other) facilities.
In a second project, funded by the DFG since June 2019, Thorn improves our basic understanding of the chemical and physical makeup of macromolecular crystals in order to improve the models used to build protein, DNA and RNA structures.
„A better understanding of the fundamental principles allows us to better interpret the data we measure in our experiments, “ says Thorn. “It allows us to solve structures where data quality and our models failed previously. In addition, with better modelling, we can potentially improve the over 100 000 already known macromolecular structures – and get more biological and medicinal answers.“ According to the structural biologist, better molecular models and higher data quality will allow scientists to fully exploit the information content of the measurements, not only in order to answer more challenging biological questions, but also to significantly improve downstream methods such as molecular dynamics and structure-based drug design.
The first prototype of AUSPEX is available on the web server of the Rudolf-Virchow Center at www.auspex.de. It will also be available to the European user community as part of the pipelines ISPyB, XDSAPP and the software suite CCP4.
Projects
„New Diagnostics for Macromolecular Structure Determination at Large Facilities: AUSPEX”
Funded by the German Federmal Ministry for Science and Education (BMBF)
Subsidy amount: 448 493 € - one postdoc position
Project start: 1st July 2019
„Towards a better understanding of macromolecular X-ray structures“
Funded by the Deutsche Forschungsgemeinschaft (DFG)
Subsidy amount: 494 722 € - Dr. Thorn’s position and one PhD student
Project start: 1st June 2019
People
Dr. Andrea Thorn is a junior group leader in the lab of Prof. Dr. Hermann Schindelin at the Rudolf-Virchow Center for Experimental Biomedicine of the University of Würzburg.
Prof. Dr. Hermann Schindelin is professor for structural biology and biochemistry and runs a research group at the Rudolf-Virchow Center for Experimental Biomedicine of the University of Würzburg since 2006.
Contact
Dr. Andrea Thorn (AG Schindelin, Rudolf Virchow Center)
Tel. +49 (0)931 31 83677, andrea.thorn@uni-wuerzburg.de
Prof. Dr. Hermann Schindelin (Rudolf Virchow Center)
Tel. +49 (0)931 31 80382, hermann.schindelin@virchow.uni-wuerzburg.de
Dr. Daniela Diefenbacher (Press Office, Rudolf Virchow Center)
Tel. +49 (0)931 31 88631, daniela.diefenbacher@uni-wuerzburg.de