P14 - Building a data-driven model of a 'virtual parasite'
Building a virtual parasite
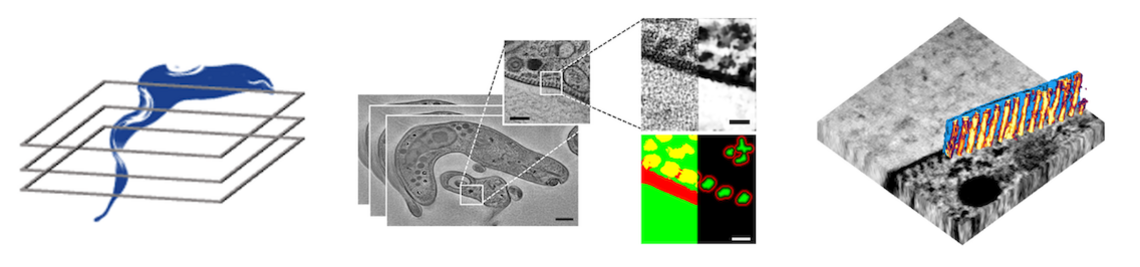
The parasitic life cycle involves a multitude of physical interactions with the host microenvironment during stages of motility and adhesion. This requires optimal adaptation of the mechanical properties of the parasite to its environment. The shape and elasticity of unicellular parasites are largely defined by their cytoskeleton. In parasitic kinetoplastids such as Trypanosoma brucei, the cytoskeleton includes a subpellicular array of microtubule filaments that forms a corset around the entire cell. How exactly the interaction between the beat of the flagellum, which is attached to the cytoskeleton and winds around the cell, and the mechanical response of the cell body, gives rise to the intricate rotational motility patterns of T. brucei is not known.
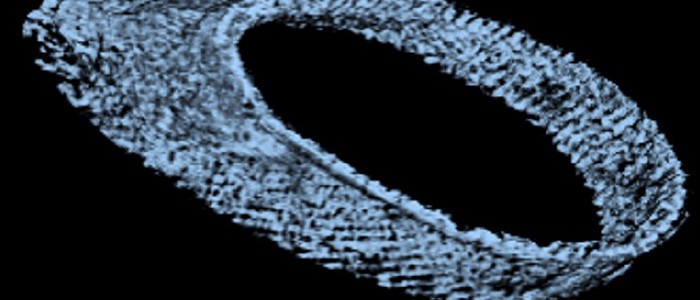
To answer this question, a detailed structural and mechanical model of the microtubule cytoskeleton and the interior of the cell, which together define its elasticity, is needed. In this project, we will build “virtual parasites” from high-resolution image data as basis for a precise data-driven mechanical understanding of parasite biophysics. In close collaboration with the group of Markus Engstler, who will provide image data and parasitology expertise, we will first perform a complete semantic segmentation (pixel-level annotation) of electron tomography image volumes of different developmental stages of T. brucei using a deep learning workflow.
First results on this objective have already been obtained and demonstrate the feasibility of our approach. We will then apply automated tracing of microtubule filaments and semi-supervised instance segmentation of the cell interior to arrive at a complete 3D structural model of the cell. This model will be converted to an annotated volume mesh amenable for finite element analysis and subjected to in-silico deformation test to validate the model against experimental data. Coarse-grained versions of the model will be generated and made available to theoretical groups studying T. brucei motility within the SPP2332. The complete workflow as well as the image data and model will be made available to all interested collaborators with via a web-based resource for visualizing and exploring the “virtual parasite”. The broader implications of building data-driven “virtual parasites” are manifold and go beyond T. brucei: by adapting the workflow to other image data, similar mechanobiological phenomena in other model parasites can be studied. Our data-driven approach will thus enable new ways of understanding the physics of parasitism by connecting imaging with computer simulations.