Research Pillar: AI for Life Science

The research pillar AI for Life Sciences aims to bridge the gap between AI and the life sciences by applying the latest AI and machine learning techniques to a wide range of research areas in the life sciences. Researchers in this pillar use cutting-edge AI technologies to analyze complex data sets, extract meaningful information, and make predictions. The goal of this research is to gain new insights and knowledge about the natural world and to improve our understanding of life science problems.
Some of the specific research areas within this pillar include Environmental Science and Ecosystems focusing on using AI methods to understand complex ecological systems and preserve biodiversity. By analyzing diverse ecological data sets and employing advanced machine learning algorithms, researchers gain a deeper understanding of ecosystems, contributing to effective conservation efforts and sustainable ecosystem management. Medical Data Analysis is another area of focus, where researchers use AI to analyze medical data and extract information that can be used for disease diagnosis, treatment planning, and population health management. In Quantitative Single-Cell Biology, researchers use AI to study the mechanisms behind cell fate decisions, and how they are controlled in a multicellular organism. The Super-Resolution research area focuses on using AI to improve the resolution of images and videos.
This research will provide new insights, knowledge, and tools that can help in the fight against diseases, environmental challenges and more.
Research Areas

The research area of "Ecosystems" is dedicated to advancing our understanding of complex ecological systems and their intricate dynamics, with a primary focus on preserving biodiversity. Ecosystems are dynamic networks of organisms and their environment, and unraveling the intricate connections and patterns within them is crucial for effective conservation and sustainable management.
Researchers in this field employ a wide range of AI methods to analyze and interpret vast and diverse ecological data sets. These datasets encompass ecological observations, satellite imagery, climate data, and other relevant sources. By applying advanced machine learning algorithms, these researchers can uncover hidden patterns, identify ecological relationships, and make accurate predictions regarding ecosystem behavior.
One of the key objectives of this research area is to develop AI-driven models that can simulate and forecast the impacts of environmental changes and human activities on ecosystems. These models provide valuable insights into how disturbances, such as climate change, habitat loss, or pollution, influence biodiversity and ecosystem services. Armed with this knowledge, scientists and policymakers can make informed decisions to conserve and restore ecosystems effectively.
Moreover, AI methods contribute to ecosystem monitoring and assessment. Through automated data collection and analysis, AI technologies can detect and monitor endangered species, track the spread of invasive species, and identify changes in vegetation cover. This enables timely interventions and proactive management strategies.
The ultimate aim of the research in the "Ecosystems" area is to foster the preservation and sustainable management of ecological systems. By leveraging AI's capabilities, researchers strive to deepen our understanding of the intricate dynamics within ecosystems and develop data-driven strategies for effective biodiversity conservation and ecosystem protection. Through interdisciplinary collaborations and cutting-edge research, this area contributes to our collective efforts in ensuring the long-term health and resilience of our planet's ecosystems.
Projects:
Principal Investigator:
Publications:
- ConvMOS: Climate Model Output Statistics with Deep Learning.
Data Mining and Knowledge Discovery, 2022.
Michael Steininger, Daniel Abel, Katrin Ziegler, Anna Krause, Heiko Paeth and Andreas Hotho.
[abstract] [BibTeX]
- Anomaly Detection in Beehives: An Algorithm Comparison.
2021.
Padraig Davidson, Michael Steininger, Florian Lautenschlager, Anna Krause and Andreas Hotho.
[BibTeX]
- DETECTING PRESENCE OF SPEECH IN ACOUSTIC DATA OBTAINED FROM BEEHIVES.
DCASE Workshop, 2021.
Pascal Janetzky, Padraig Davidson, Michael Steininger, Anna Krause and Andreas Hotho.
[BibTeX]
- Evaluating the multi-task learning approach for land use regression modelling of air pollution.
In: International Conference on Frontiers of Artificial Intelligence and Machine Learning. 2020.
Andrzej Dulny, Michael Steininger, Florian Lautenschlager, Anna Krause and Andreas Hotho.
[abstract] [BibTeX]
- OpenLUR: Off-the-shelf air pollution modeling with open features and machine learning.
Atmospheric Environment, 233:117535, 2020.
Florian Lautenschlager, Martin Becker, Konstantin Kobs, Michael Steininger, Padraig Davidson, Anna Krause and Andreas Hotho.
[doi] [abstract] [BibTeX]
- MapLUR: Exploring a new Paradigm for Estimating Air Pollution using Deep Learning on Map Images.
2020. cite arxiv:2002.07493.
Michael Steininger, Konstantin Kobs, Albin Zehe, Florian Lautenschlager, Martin Becker and Andreas Hotho.
[doi] [abstract] [BibTeX]
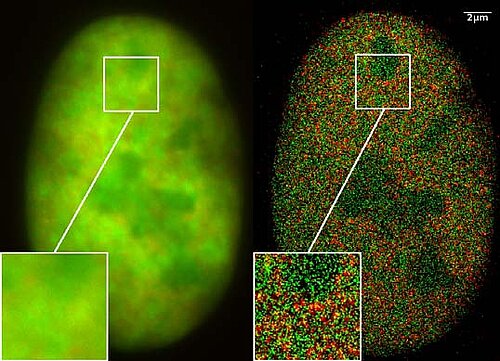
Super-resolution is a technique used in computer vision to increase the resolution of an image or video, while also improving its visual quality. The goal of super-resolution is to take a low-resolution image or video and produce a high-resolution version that is more detailed and visually pleasing. CAIDAS researchers have already made significant contributions in this field, specifically in the area of Single-Image Super-Resolution (SISR) which aims to improve the resolution of a single image by using a model trained on a large dataset of high-resolution images. The goal of this research is to develop and improve upon the current state-of-the-art algorithms and models for super-resolution, and to apply these techniques to real-world problems such as image and video processing, computer vision and more.
Principal Investigator:
Publications:
- SRFlow: Learning the Super-Resolution Space with Normalizing Flow..
In: A. Vedaldi, H. Bischof, T. Brox and J.-M. Frahm, editors, ECCV (5), volume 12350, series Lecture Notes in Computer Science, pages 715-732. Springer, 2020.
Andreas Lugmayr, Martin Danelljan, Luc Van Gool and Radu Timofte.
[doi] [BibTeX]
Cell fate decision is essential for the emergence of a complex multicellular organism starting from a single pluripotent zygote, and perpetually occurs during adult life to maintain organ tissues. The robustness of cell fate decision is remarkable given the stochastic interaction of hundreds of thousands of molecules in each cell. It has remained a central open question how signals from the microenvironment are integrated with stochastic processes in a cell to control cell fate decision in normal tissues and upon perturbations due to disease or tissue damage. Our lab investigates these questions in the context of immune cell differentiation by combining single-cell resolution experimental methods with computational methods involving machine learning and mathematical modeling.
Projects:
- TissueNet
- Deciphering cell fate decision at single-cell resolution
- Regulation of gene expression during cellular differentiation
- The role of biological gene expression noise during cellular differentiation
- Differentiation of immune cells
- Cell differentiation and plasticity in the healthy and diseased liver
Principal Investigator:
- Dominic Grün
- Andreas Hotho Ingo Scholtes
- Ingo Scholtes
Publications:
- Developmental dynamics of two bipotent thymic epithelial progenitor types.
Nature, 606(7912):165-171, 2022.
Anja Nusser, Sagar, Jeremy B. Swann, Brigitte Krauth, Dagmar Diekhoff, Lesly Calderon, Christiane Happe, Dominic Grün and Thomas Boehm.
[doi] [BibTeX]
- Deciphering Cell Fate Decision by Integrated Single-Cell Sequencing Analysis.
Annual Review of Biomedical Data Science, 3(1):1-22, 2020.
Sagar and Dominic Grün.
[doi] [BibTeX]
- In Situ Maturation and Tissue Adaptation of Type 2 Innate Lymphoid Cell Progenitors.
Immunity, 53(4):775-792.e9, 2020.
Patrice Zeis, Mi Lian, Xiying Fan, Josip S. Herman, Daniela C. Hernandez, Rebecca Gentek, Shlomo Elias, Cornelia Symowski, Konrad Knöpper, Nina Peltokangas, Christin Friedrich, Remi Doucet-Ladeveze, Agnieszka M. Kabat, Richard M. Locksley, David Voehringer, Marc Bajenoff, Alexander Y. Rudensky, Chiara Romagnani, Dominic Grün and Georg Gasteiger.
[doi] [BibTeX]
- A human liver cell atlas reveals heterogeneity and epithelial progenitors.
Nature, 572(7768):199-204, 2019.
Nadim Aizarani, Antonio Saviano, Sagar, Laurent Mailly, Sarah Durand, Josip S. Herman, Patrick Pessaux, Thomas F. Baumert and Dominic Grün.
[doi] [BibTeX]
CAIDAS works with environmental data to contribute to the ongoing climate and ecosystem research. Based on low-cost sensors, map and geographical data, we develop novel algorithms to address environmental questions. These questions include "What is the air pollution at a given spot on earth?" and "What type of wine should I grow on my land considering climate change in the next few decades?". While answering such questions is one concern of our research, the other objective is to raise awareness for the environment in the society.
At the intersection of AI and climate research, we target the use of advanced machine learning for climate models.
Bees are an important part of our ecosystem and we focus our research on analyzing data collected from smart beehives using machine learning approaches. Typical research questions are: Can we automatically detect swarms without labels? What is the current state of the bee colony? Will my bee colonies survive the winter?
Projects:
Principal Investigator:
Publications:
- ConvMOS: Climate Model Output Statistics with Deep Learning.
Data Mining and Knowledge Discovery, 2022.
Michael Steininger, Daniel Abel, Katrin Ziegler, Anna Krause, Heiko Paeth and Andreas Hotho.
[abstract] [BibTeX]
- Anomaly Detection in Beehives: An Algorithm Comparison.
2021.
Padraig Davidson, Michael Steininger, Florian Lautenschlager, Anna Krause and Andreas Hotho.
[BibTeX]
- DETECTING PRESENCE OF SPEECH IN ACOUSTIC DATA OBTAINED FROM BEEHIVES.
DCASE Workshop, 2021.
Pascal Janetzky, Padraig Davidson, Michael Steininger, Anna Krause and Andreas Hotho.
[BibTeX]
- Semi-Supervised Learning for Grain Size Distribution Interpolation.
In: Pattern Recognition. ICPR International Workshops and Challenges: Virtual Event, January 10-15, 2021, Proceedings, Part VI, pages 34-44. 2021.
Konstantin Kobs, Christian Schäfer, Michael Steininger, Anna Krause, Roland Baumhauer, Heiko Paeth and Andreas Hotho.
[BibTeX]
- Density-based weighting for imbalanced regression.
Machine Learning, 110(8):2187-2211, 2021.
Michael Steininger, Konstantin Kobs, Padraig Davidson, Anna Krause and Andreas Hotho.
[doi] [abstract] [BibTeX]
- Evaluating the multi-task learning approach for land use regression modelling of air pollution.
In: International Conference on Frontiers of Artificial Intelligence and Machine Learning. 2020.
Andrzej Dulny, Michael Steininger, Florian Lautenschlager, Anna Krause and Andreas Hotho.
[abstract] [BibTeX]
- OpenLUR: Off-the-shelf air pollution modeling with open features and machine learning.
Atmospheric Environment, 233:117535, 2020.
Florian Lautenschlager, Martin Becker, Konstantin Kobs, Michael Steininger, Padraig Davidson, Anna Krause and Andreas Hotho.
[doi] [abstract] [BibTeX]
- MapLUR: Exploring a new Paradigm for Estimating Air Pollution using Deep Learning on Map Images.
2020. cite arxiv:2002.07493.
Michael Steininger, Konstantin Kobs, Albin Zehe, Florian Lautenschlager, Martin Becker and Andreas Hotho.
[doi] [abstract] [BibTeX]
- MixedTrails: Bayesian hypothesis comparison on heterogeneous sequential data.
Data Mining and Knowledge Discovery, 2017.
Martin Becker, Florian Lemmerich, Philipp Singer, Markus Strohmaier and Andreas Hotho.
[doi] [abstract] [BibTeX]
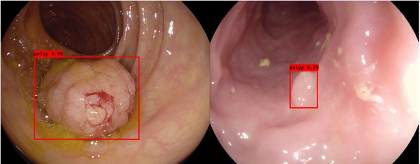
Medical Data Analytics is a rapidly growing field that uses advanced analytical and statistical techniques to extract meaningful insights from large amounts of medical data. This data can come from a variety of sources such as electronic health records, medical imaging, clinical trials, and wearable devices. The goal of Medical Data Analytics is to improve patient care and outcomes by identifying patterns and trends that can inform diagnosis, treatment, and disease management. The use of machine learning and artificial intelligence algorithms is becoming increasingly prevalent in this field, allowing for the detection of complex patterns and relationships in data that may not be visible to the human eye. Applications of Medical Data Analytics include, but are not limited to, disease diagnosis and prognosis, drug development and personalized medicine, clinical decision support and population health management. The field continues to evolve as new technologies and data sources become available, offering new opportunities for medical research and improving patient care.
Projects:
Principial Investigators:
Publications:
- IT-Supported Request Management for Clinical Radiology: Contextual Design and Remote Prototype Testing.
In: CHI Conference on Human Factors in Computing Systems Extended Abstracts, series CHI EA '22, pages 1-8. Association for Computing Machinery, New York, NY, USA, 2022.
Philipp Krop, Samantha Straka, Melanie Ullrich, Maximilian Ertl and Marc Erich Latoschik.
[doi] [abstract] [BibTeX]
- Towards Responsible Medical Diagnostics Recommendation Systems.
2022. cite arxiv:2209.03760Comment: 4th FAccTRec Workshop on Responsible Recommendation.
Daniel Schlör and Andreas Hotho.
[doi] [abstract] [BibTeX]
- Fast Machine Learning Annotation in the Medical Domain: A Semi-Automated Video Annotation Tool for Gastroenterologists.
BioMedical Engineering OnLine volume, 21(33), 2021.
Adrian Krenzer, Kevin Makowski, Amar Hekalo, Daniel Fitting, Joel Troya, Wolfram G Zoller, Alexander Hann and Frank Puppe.
[doi] [BibTeX]
Principial Investigators
Radu Timofte
Computer Vision
Dominic Grün
Computational Biology of Spatial Biomedical Systems
Andreas Hotho
Data Science
Frank Puppe
Artificial Intelligence and Knowledge Systems
Ingo Scholtes
Machine Learning for
Complex Networks
N.N.
Artificial Intelligence for the Molecular Sciences
N.N.
AI in Computational and Theoretical Biology
N.N.
Pattern Recognition