Johannes Balkenhol
I am a researcher specializing in investigating biological systems using network models, and I play a pivotal role in communicating the tenets of research data management.
My primary research focus is on modeling biological systems to study dynamics both in vivo and in silico. The process involves the following key modeling steps: setting up a model, observing its behavior, modifying the behavior, and continuing the modified model. These models are constantly refined using mathematical, physical, and biological data to ensure they are tailored to specific use cases.
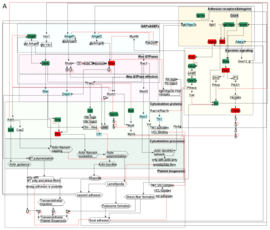
In particular, I integrate experimental data such as omics, imaging, and interaction data to reconstruct network models. In bioinformatics, we use algorithms to simulate the models dynamically over time and space. The subsequent simulated system behavior is then validated through experiments. As use cases, I studied the dynamic behavior of platelets, which are key players in hemostasis and diseases such as stroke and heart attack. Furthermore, I tried to unravel certain processes involved in fungal immune evasion and studied oscillatory networks to model cortical information processing.
By comparing the different biological network topologies and dynamics, I aim to uncover fundamental principles of biological processing and trace an evolutionary path that is potentially indicated by maximizing information integration.
The high integration of big data from different research branches forces the establishment of a “good” research data management (RDM) system. The ever-increasing size of imaging and omics data, in particular, necessitates superior data management methodologies.
Therefore, as a member of the IZKF core unit Research Data Management at the Center for Integrative and Translational Bioimaging, I am responsible for the communication and onboarding of the Use Cases (collaborative research centers) of the Virtual Research Campus (VRC). Careful RDM using the FAIR principles facilitates data exchange, maintenance, integration, and long-term usage.
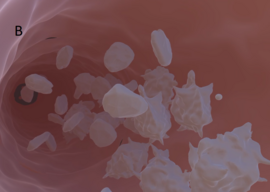
As a continuation of “good” RDM, we establish a virtual 3D interactive platelet simulation - a collaborative project of the Heinze Lab, Bioinformatics, and Chair of Games Engineering Würzburg. This multidisciplinary research data hub includes data from different scientific fields, such as clinical and biological platelet research, bioinformatics, imaging, and game engineering. Our vision is for this integrative platform to serve as a virtual research environment and knowledge repository for platelet researchers and a navigable space for both novices and experts.