P 8: J. Vogel / D. Zdzieblo
Establishing a human intestinal tissue model to study host and pathogen determinants that restrict Salmonella enterica infection
State of the art
Salmonella bacteria constitute some of the most well studied human pathogens. Of these, primate-restricted subspecies Salmonella enterica serovar Typhi (S. Typhi) causes life-threatening typhoid fever, while S. Typhimurium has a broad host-range and causes non-typhoidal intestinal infections and enteritis in humans. Both infect their hosts oraly, exhibit a facultative intracellular life-style, and extensively manipulate host immune signaling [1]. While S. Typhimurium constitutes one of the best studied laboratory models for enteric bacterial infections, S. Typhi interactions with human host cells remain little understood due to the limited availability of primate infection models. Moreover, small animal infection models of S. Typhimurium infection do not accurately reflect the innate immune response of the human host due to differences in disease progression and pattern recognition [2, 3]. Infection models based on engineered human tissue might help to overcome those limitations.
Previous Work
We have begun to establish 3D in vitro human infection models based on Caco-2 and primary intestinal cells mimicking the gastrointestinal barrier. In a first approach, we used static transwell models based on an acellularized matrix of porcine gut fixed in a cell-crown. The apical side of the matrix is colonized with a monolayer of differentiated, polarized Caco-2 cells; the basolateral side with primary human endothelial cells [4]. The apical epithelial surface represents the luminal side of the gut and the basolateral side the vascular system. Thus far, we have challenged this tissue model with S. Typhimurium, in preparation for infections with S. Typhi.
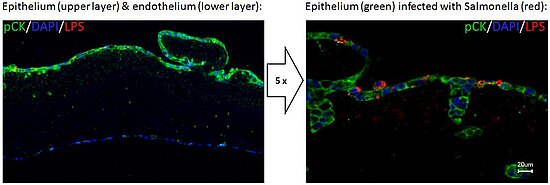
Figure: Microscopic immunofluorescence images of a cross-section of an intestinal barrier model infected with S. Typhimurium. pCK = epithelial cell marker; DAPI: nuclei stain; LPS: S. Typhimurium lipopolysacharide.
Our preliminary results with GFP-positive S. Typhimurium applied to the apical compartment followed by flow cytometry demonstrate robust infection of the epithelial cell layer (20-30 % infected cells after 24 hours at an MOI of 10). In agreement with the known restriction of S. Typhimurium infection to the human gut epithelium no dissemination into the subjacent endothelial cell layer could be detected. We have however found evidence of a low-level transmission of the bacteria across the intestinal barrier into the vascular medium which might explain systemic S. Typhimurium infestations in immune-compromised patients [5].
To create an infection model closely mimicking the immune reactions occurring in native human tissue we have run a pilot experiment with the above described static tissue models colonized with human peripheral blood mononuclear cells (PBMCs). To this end, a PBMC preparation was added to the basolateral compartment mimicking the vascular system. To demonstrate feasibility of transcriptional profiling in multiple major components of the host immune system and the pathogen simultaneously, we purified defined cell populations of the tissue model by cell sorting. Twenty-four hours post infection with GFP-positive S. Typhimurium, the tissue was disrupted by trypsinization and bystander and GFP+ epithelial cells, endothelial cells, CD14+ monocytes CD56+ NK-cells and CD4+ T-cells were purified. Sufficient amounts of RNA (> 500 ng) required for transcriptional profiling by RNA-Seq were obtained from all cellular fractions.
Work Plan
Next, we plan to use advanced intestinal tissue models using primary human epithelial cells and apply a constant flow of medium in the apical chamber and pulsatile flow in the vascular compartment within a bioreactor. Cells cultured under these conditions show more in vivo-like characteristics as judged by morphology, barrier function and protein expression. Differentiated primary cells represent a variety of cell types such as enterocytes, mucus-producing goblet cells and enteroendocrine cells, which all contribute interactions between host and pathogen. The vascular compartment will be supplemented with purified PBMC preparations to understand the role of these cells in human Salmonella infection. Using dual RNA-Seq [6] we will simultaneously resolve human and S. Typhi transcriptomic changes in all cellular components of the PBMC-supplemented human tissue models as the infection progresses. In continuation with our previous work [7] the emphasis will be on the discovery of non-coding RNA functions in the host immune response and the pathogen counter-response. The use of engineered human tissue models will allow us to study the role of ncRNA in the interplay of human cells and host-adapted Salmonella with unprecedented resolution.
References
- Raymond et al. (2013) Subversion of Trafficking, Apoptosis, and Innate Immunity by Type III Secretion System Effectors. Trends in Microbiology 21(8):430-441. PubMed
- Salzman et al. (2003) Protection against enteric salmonellosis in transgenic mice expression a human intestinal defensin. Nature 422, 522-526. PubMed
- Mathur et al. (2012) A mouse model of Salmonella typhi infection, Cell 151(3):590-602. PubMed
- Pusch et al. (2011) The physiological performance of a three-dimensional model that mimics the microenvironment of the small intestine. Biomaterials 32(30):7469-78. PubMed
- Graham SM (2010) Nontyphoidal Salmonellosis in Africa. Curr Opin Infect Dis 23:409-414. PubMed
- Westermann et al. (2012) Dual RNA Seq of pathogen and host. Nature Reviews Microbiology 10:618-630. PubMed
- Westermann et al. (2016) Dual RNA-Seq unveils non-coding RNA functions in Salmonella-host interplay. Nature. 529:496-501. PubMed